|
 |
|
Microarrays allow gene expression levels to be quantified for all genes
in a particular cell type of an organism.
By analysing such data, we can find out which genes are expressed
at particular developmental stages or under particular
environmental conditions (for
example, when a drug is present or absent); or compare expression
in different cell types (for example, normal and cancerous cells).
|  |
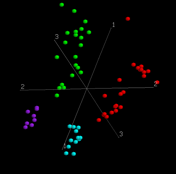
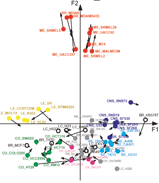
We are interested in the application of multivariate statistical methods to the analysis of microarray gene expression and proteomics data. Recent work has focused on classification and prediction of cancer classes using Between Group Analysis, cross-platform analysis of array data using CoInertia Analysis, and correlation of gene expression data with other sources of data, including clinical, pharmacological and proteomic data.
Our lab is collaborating with Guy Perriere, Lyon in using multivariate statistical methods to analyse cancer data sets. We are also collaborating with Professor Gerry O'Sullivan and colleagues at the Cork Cancer Research Centre to analyse data from oesophageal cancer and to set up a microarray analysis facility in Cork. We also collaborate with Dr. Liam Gallagher's Lab and other Labs in the Conway Institute, UCD.
For more information on our research into microarray data analysis,
please see
- our website on Between Group Eigen Analysis of microarray data, which accompanies our paper Culhane, A. C., Perriere, G., Considine, E. C., Cotter, T. G. and Higgins, D. G. (2002) Between-group analysis of microarray data. Bioinformatics. 18:1600-1608.
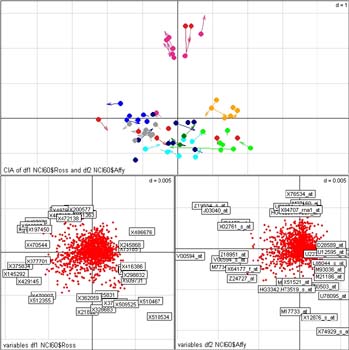
- our website on Cross platform analysis of microarray Datasets using coinertia analysis, which accompanies our paper Culhane, A. C., Perriere, G. and Higgins, D. G. (2003) Cross-platform comparison and visualisation of gene expression data using co-inertia analysis. BMC Bioinformatics. 4(1):59.
- Our website on our R package for multivariate analysis of microarray data MADE4
- Aedín's home page